FROGS News - June 2023
FROGS v4.1.0 is available
FROGS v4.1.0 can be downloaded on  :
:
https://github.com/geraldinepascal/FROGS/releases
https://github.com/geraldinepascal/FROGS-wrappers/releases
For an easy installation there are :
 : https://anaconda.org/bioconda/frogs
: https://anaconda.org/bioconda/frogs
 : https://toolshed.g2.bx.psu.edu/view/frogs/frogs/37e6f0c959bb
: https://toolshed.g2.bx.psu.edu/view/frogs/frogs/37e6f0c959bb
This new version is available among others on
What has changed since the last version?
FROGS produces ASV:
“ASVs are identical denoised reads with as few as 1 base pair difference between variants, representing an inference of the biological sequences prior to amplification and sequencing errors (Callahan et al., 2017). ASVs are mainly formed through the two most popular denoising algorithms, DADA2 and UNOISE (Edgar, 2016). Although less frequently implemented in the pipeline, deblur (Amir et al. 2017) and obiclean (Boyer et al., 2016) are other denoising algorithms for ASVs formation. The OTU clustering approaches include a much wider set of algorithms across different software, which typically rely on global sequence similarities. Swarm (Mahe et al., 2022) is a notably different sequence clustering approach, which, while technically a clustering algorithm, may also be considered a denoising method when using the fastidious method with d=1. It relies on the maximum number of differences between reads (local linking threshold) and forms clusters that are resilient to input-order changes, thus creating stable, high-resolution features (herein referred to as swarm-clusters). When using the fastidious method with d=1, swarm aims to produce clusters centered around real biological sequences, where clusters represent sequence variants.”"
Since FROGS uses swarm (with the fastidious method with d=1) and strongly promotes denoising by chimera removal and cluster filtering, FROGS produces ASVs.
As a result, we have modified all the terminology and changed two tool names:
- FROGS OTU Filters -> FROGS_4 Cluster filters
- FROGS Affiliation OTU -> FROGS_5 Taxonomic affiliation
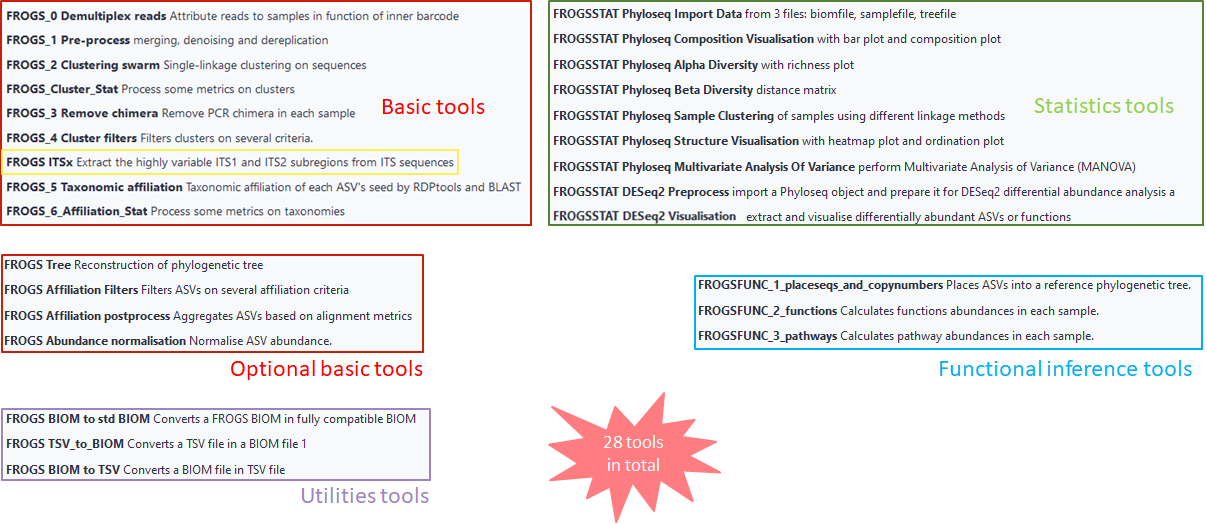
More readability on the 7 basic steps
For more readability, now the 7 basic steps are named:
- FROGS_0 Demultiplex reads: attributes reads to samples in function of inner barcode
- FROGS_1 Pre-process: merging, denoising and dereplication
- FROGS_2 Clustering swarm: produces single-linkage clustering on sequences
- FROGS_3 Remove chimera: removes PCR chimera in each sample
- FROGS_4 Cluster filters: filters clusters on several criteria
- FROGS_5 Taxonomic affiliation: produces taxonomic affiliation of each ASV’s seed by RDPtools and BLAST
- FROGS_6_Affiliation_Stat: processes some metrics on taxonomies
FROGS_1 Pre-process:
FROGS_1 Pre-process filters and dereplicates amplicons for use in diversity analysis. Now, this tool takes in charge long reads, you can choose between:
- short reads : Illumnia Miseq , Hiseq (paired-ends or single-ends)
- long reads : PACBIO or Oxford Nanopore Technology (single-ends)
- short reads : 454 (single-ends)
Functional inference:
The 4 FROGSFUNC tools are now 3 FROGSFUNC tools
- FROGSFUNC_1_placeseqs_and_copynumbers: places ASVs into a reference phylogenetic tree.
- FROGSFUNC_2_functions: calculates functions abundances in each sample.
- FROGSFUNC_3_pathways: calculates pathway abundances in each sample.
Also, we fixed some bugs and improve the tools and their outputs. We have added companion graphics to help with the decision making process.
Differential analysis:
The package DESeq2 provides methods to test for differential expression by use of negative binomial generalized linear models
- 1st step: FROGSSTAT DESeq2 Preprocess
- 2nd step : FROGSSTAT DESeq2 Visualisation
Now,  these two tools are available to analyse ASV or Function (from FROGSFUNC tools).
these two tools are available to analyse ASV or Function (from FROGSFUNC tools).
Affiliation filter:
This tool removes or keeps ASVs or hides taxonomical metadata according to one or more criteria:
- for RDP taxonomy : a minimal bootstrap threshold at a specific rank
- for blast taxonomy : a minimal identity rate, coverage rate, or alignment length, or a maximal evalue, or the absence/presence of a full or partial taxon name.
Now, user can choose to filter based on affiliation keyword in keeping all ASVs with the keyword. Before, user could only ignore ASVs with the keyword.
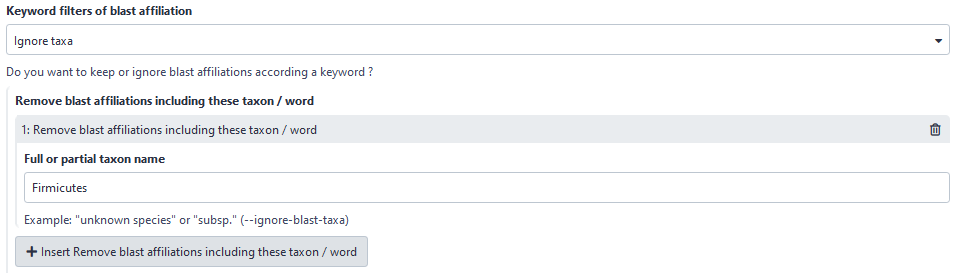
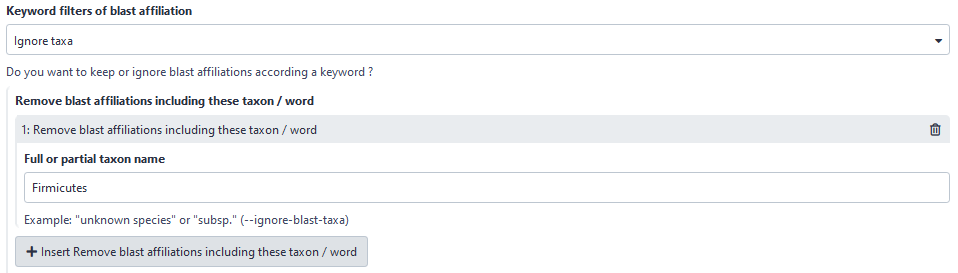
Here, user will ignore all ASVs with “Firmicutes” in its taxonomic affiliation.
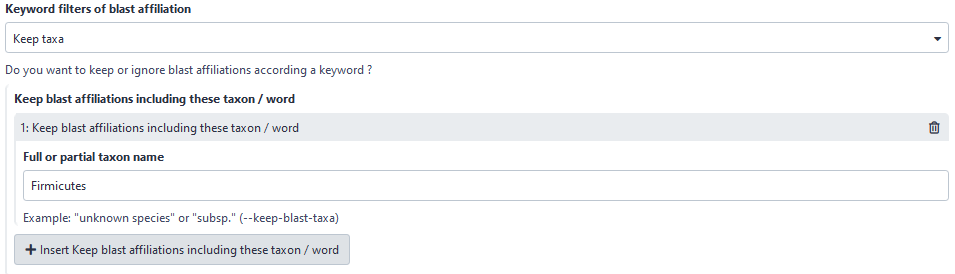
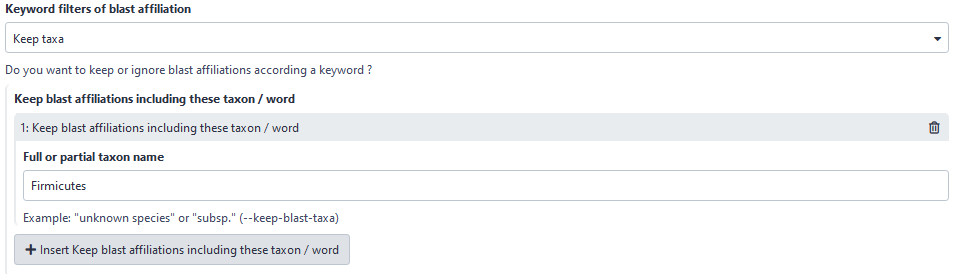
Here, user will keep all ASVs with “Firmicutes” in its taxonomic affiliation.
New documentations for using FROGS v4.1.0:
New databases are available
http://genoweb.toulouse.inra.fr/frogs_databanks/assignation/readme.txt, here are all the databases we have formatted (on demand) for RDPClassifier and NCBI Blast+
Please pay attention to the licence of the database and how to cite it.
| amplicon |
base |
filters |
version |
galaxy_value |
| ITS |
UNITE |
Eukaryote |
9.0 |
ITS_UNITE_Eukayote_9.0 |
| ITS |
UNITE |
Fungi |
9.0 |
ITS_UNITE_Fungi_9.0 |
| 23S |
microgreen-db |
algae |
1.2 |
23S_microgreen-db_algae_v1.2 |
| 23S |
microgreen-db |
ncbi |
1.2 |
23S_microgreen-db_ncbi_v1.2 |
| 23S |
microgreen-db |
pr2-silva |
1.2 |
23S_microgreen-db_pr2-silva_v1.2 |
| rbcL |
Diat.barcode |
|
11.1 |
rbcL_Diat.barcode_11.1 |
| 18S |
Diat.barcode |
|
11.1 |
18S_Diat.barcode_11.1 |
| COI |
COInr |
|
2022_05_06 |
COI_COInr_2022_05_06 |
| COI |
MIDORI2 |
UNIQ_SP |
GB253 |
COI_MIDORI2_UNIQ_SP_GB253 |
| COI |
MIDORI2 |
LONGEST_SP |
GB253 |
COI_MIDORI2_LONGEST_SP_GB253 |
You need help to use FROGS, you are looking for training:
Please contact frogs-support@inrae.fr
And/or visit:
for a webinar training: http://frogs.toulouse.inrae.fr/html/training.php
 next session: 9th to 12th October 2023
next session: 9th to 12th October 2023
for a presential training at Jouy-en-Josas:
https://migale.inrae.fr/trainings
 next session: 10th to 14th September 2023
next session: 10th to 14th September 2023
Who uses FROGS?
FROGS News - June 2023
FROGS News - June 2023
FROGS v4.1.0 is available
FROGS v4.1.0 can be downloaded on :
:
https://github.com/geraldinepascal/FROGS/releases
https://github.com/geraldinepascal/FROGS-wrappers/releases
For an easy installation there are :
 : https://anaconda.org/bioconda/frogs
: https://anaconda.org/bioconda/frogs
 : https://toolshed.g2.bx.psu.edu/view/frogs/frogs/37e6f0c959bb
: https://toolshed.g2.bx.psu.edu/view/frogs/frogs/37e6f0c959bb
This new version is available among others on
What has changed since the last version?
FROGS produces ASV:
“ASVs are identical denoised reads with as few as 1 base pair difference between variants, representing an inference of the biological sequences prior to amplification and sequencing errors (Callahan et al., 2017). ASVs are mainly formed through the two most popular denoising algorithms, DADA2 and UNOISE (Edgar, 2016). Although less frequently implemented in the pipeline, deblur (Amir et al. 2017) and obiclean (Boyer et al., 2016) are other denoising algorithms for ASVs formation. The OTU clustering approaches include a much wider set of algorithms across different software, which typically rely on global sequence similarities. Swarm (Mahe et al., 2022) is a notably different sequence clustering approach, which, while technically a clustering algorithm, may also be considered a denoising method when using the fastidious method with d=1. It relies on the maximum number of differences between reads (local linking threshold) and forms clusters that are resilient to input-order changes, thus creating stable, high-resolution features (herein referred to as swarm-clusters). When using the fastidious method with d=1, swarm aims to produce clusters centered around real biological sequences, where clusters represent sequence variants.”"
Since FROGS uses swarm (with the fastidious method with d=1) and strongly promotes denoising by chimera removal and cluster filtering, FROGS produces ASVs.
As a result, we have modified all the terminology and changed two tool names:
Modified tools:
More readability on the 7 basic steps
For more readability, now the 7 basic steps are named:
FROGS_1 Pre-process:
FROGS_1 Pre-process filters and dereplicates amplicons for use in diversity analysis. Now, this tool takes in charge long reads, you can choose between:
Functional inference:
The 4 FROGSFUNC tools are now 3 FROGSFUNC tools
Also, we fixed some bugs and improve the tools and their outputs. We have added companion graphics to help with the decision making process.
Differential analysis:
The package DESeq2 provides methods to test for differential expression by use of negative binomial generalized linear models
Now, these two tools are available to analyse ASV or Function (from FROGSFUNC tools).
these two tools are available to analyse ASV or Function (from FROGSFUNC tools).
Affiliation filter:
This tool removes or keeps ASVs or hides taxonomical metadata according to one or more criteria:
Now, user can choose to filter based on affiliation keyword in keeping all ASVs with the keyword. Before, user could only ignore ASVs with the keyword.
Here, user will ignore all ASVs with “Firmicutes” in its taxonomic affiliation.
Here, user will keep all ASVs with “Firmicutes” in its taxonomic affiliation.
New documentations for using FROGS v4.1.0:
To try FROGS on Galaxy server (documentation + datasets):
http://genoweb.toulouse.inra.fr/~formation/15_FROGS/current/
Tutorials are now availbale to use FROGS on Galaxy and in command line:
 Come back to visit this page regularly, the tutorials will grow over the months.
Come back to visit this page regularly, the tutorials will grow over the months.
http://frogs.toulouse.inrae.fr/html/tuto.html
New databases are available
http://genoweb.toulouse.inra.fr/frogs_databanks/assignation/readme.txt, here are all the databases we have formatted (on demand) for RDPClassifier and NCBI Blast+
Please pay attention to the licence of the database and how to cite it.
You need help to use FROGS, you are looking for training:
Please contact frogs-support@inrae.fr
And/or visit:
 next session: 9th to 12th October 2023
next session: 9th to 12th October 2023
for a webinar training: http://frogs.toulouse.inrae.fr/html/training.php
for a presential training at Jouy-en-Josas:
 next session: 10th to 14th September 2023
next session: 10th to 14th September 2023
https://migale.inrae.fr/trainings
Who uses FROGS?
A work by FROGS team